Methane-Cycling Microbiome in Permafrost
Summary:
- Taxonomic and functional profiling of methane cycling microbiomes in permafrost soil
- Characterization of microbial diversity via 16S rRNA amplicon data anlysis
- Evaluation of active microbial populations by targeting microorganisms contributing to greenhouse gas (CH4 and CO2) production
Project Topic:
Analysis of methane-cycling microbiome in permafrost
Project Partner:
GFZ Potsdam, UiT, CAREERI
Location:
Siberia, Svalbard, Qinghai-Tibet Plateau
Sample type:
Permafrost soils
Target Molecule:
genomic DNA
Laboratory Analysis:
16S rRNA gene and metagenomic sequencing (Illumina) of genomic DNA retrieved from the active layer and frozen permafrost, as well as thermokarst lakes and lagoons which are subject to rapid thawing.
Bioinformatic Analysis:
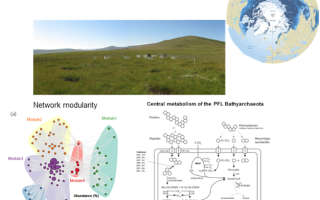
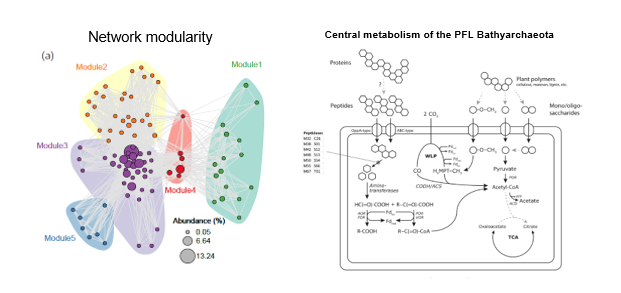
Quality control, demultiplexing, taxonomic classification, metagenome assembly and binning, genome quality assessment, functional annotation, metabolic pathway reconstruction, downstream analysis including network analysis, LEfSe, ordination, SEM and multiple other numeric analyses.
Summary:
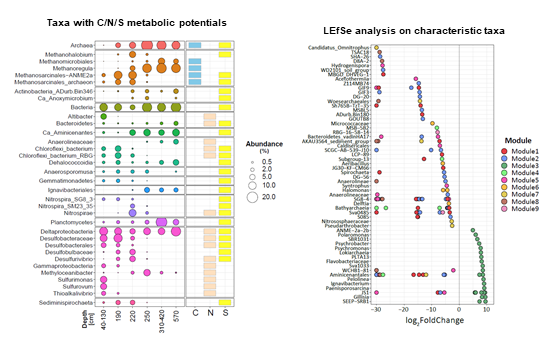
Taxonomic and functional profiling revealed high diversity of methane cycling microbiome residing in permafrost soil, with heterogeneous distribution patterns in active layer and permafrost.
More details:
https://doi.org/10.1111/mec.16118
https://doi.org/10.1002/ppp.2131
https://doi.org/10.1016/j.soilbio.2017.03.007