GREENGATE GENOMICS
SERVICES
Microbial analysis
service
Do you want to know about the microorganisms in your samples. Learn more about different options to get the best solution for you.
Bioinformatic
service
Bioinformatics of sequenencing data is our passion. Get in touch to see how we can help!
Specific SERVICES
Amplicon
service
From amplicon PCR in the lab, over low cost sequencing to high quality analysis. We are the perfect partner for your amplicon project!
Metagenomic
service
Do you need metagenomic sequencing or analysis? We are your partner for any metagenomics application!
Custom software
and service
Plan your experiment to optimize sequencing cost and analysis quality? Do you need custom bioinformatic service or software? Let’s discuss the details!
ABOUT
GREENGATE GENOMICS
GreenGate Genomics is a cutting-edge bioinformatic service startup specializing in microbial and environmental genomics. Founded in 2023 by five researchers at the GFZ German Research Center for Geosciences, we are at the forefront of deciphering the world of microbial genetic data. With a team of skilled bioinformaticians and molecular microbiologists, we utilize advanced, state-of-the-art techniques and workflows to provide comprehensive analysis and interpretation of microbial genomes. Our tailored services cater to the unique needs of academic researchers, biotech companies, and healthcare professionals, equipping them with invaluable insights into the complexities of microbial systems.
GREENGATE GENOMICS
CASE STUDIES
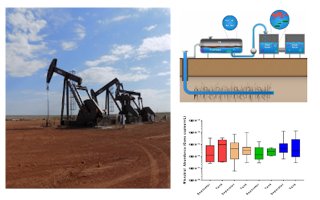
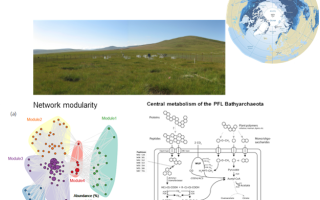
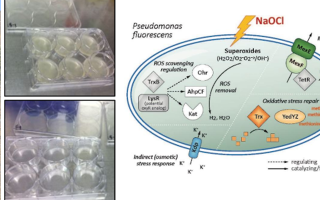
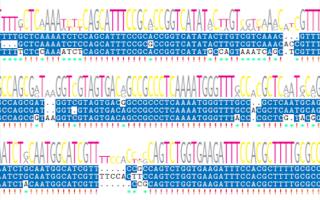
We at GreenGate Genomics have extensive expertise working in the field of environmental microbial genomics. Some of the projects we have participated in are listed below. Our work has also been published in numerous peer reviewed research articles in renowned scientific journal. Please have a look at our publications page.