
Oil and Gas Ecology
Evaluation of microbial abundance and composition patterns highlighted
elevated potential for biocorrosion and well fouling. Statistical assessment
suggested a link between biocide usage and microbial
We at GreenGate Genomics have extensive expertise working in the field of environmental microbial genomics. Some of the projects we have participated in are listed below. Our work has also been published in numerous peer reviewed research articles in renowned scientific journal. Please have a look at our publications page.

Evaluation of microbial abundance and composition patterns highlighted
elevated potential for biocorrosion and well fouling. Statistical assessment
suggested a link between biocide usage and microbial
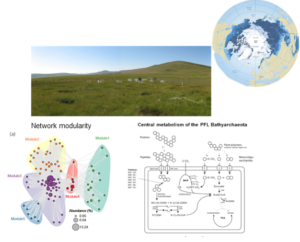
Taxonomic and functional profiling revealed high diversity of methane cycling microbiome residing in permafrost soil, with heterogeneous distribution patterns in active layer and permafrost. Diversity

Specifically adapted laboratory and bioinformatic workflows allowed the reconstruction of microbial distribution patterns down to a depth of 230 m across this, difficult to work
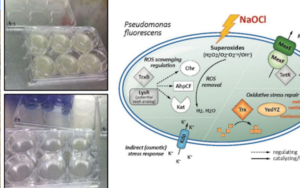
Differential expression analysis resulted in the identification of significantly upand downregulated genes in response to sodium hypochlorite. Following annotation of these genes, results were used

Evaluating the extracellular and intracellular DNA fractions in soil and rock samples from different Atacama desert locations, it was possible to identify highly adapted microbial
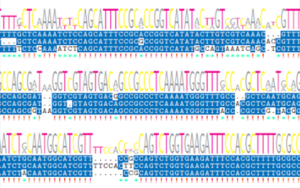
Oxford Nanopore Technologies (ONT) offers long read sequencing by nanopore sequencing. Long reads are well suited for genome reconstruction, still the workflow requires careful execution
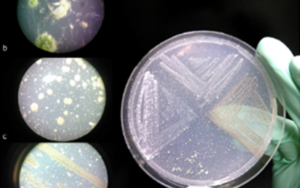
Six different bacterial strains were isolated from plastic polluted landfill soil, taxonomically classified and their genomes sequenced and reconstructed. Functional Annotation allowed the discovery of
© 2023 greengategenomics.com